LARA and NFDI4Cat -
how can research data be automatically semantically annotated?
mark doerr & stefan maak & KIWI / SiLA / AnIML / NFDI4Cat teams institute for biochemistry, university
greifswald
Kiel, 2024-05-12


Any element with the class="notes" will not be displayed. This can
be used for speaker notes. In fact, the impressConsole plugin will
show it in the speaker console!
Press ctrl-C to activate the console

NFDI4Cat Vocabularies
NFDI4Cat-TA1, David Linke, Nikoloas Moustakas
source: https://github.com/nfdi4cat/voc4cat
*

ontology world map
NFDI4Cat-TA1, Hendrik Borgelt, Alexander Behr
source: https://github.com/nfdi4cat/Ontology-Overview-of-NFDI4Cat
*

PID service
NFDI4Cat-TA1, David Linke, Preston Rodrigues
- unique identifier for all data series, device, etc.
- handle based Persistent Identifier (PID) Service
- demo setup at the HLRS Stuttgart
*

metadata standards, local pilot installations,
central services
NFDI4Cat-TA2 / TA3 / TA4
- metadata standards for (bio-)catalytic experiments - JSON-LD / DCAT-AP
- local pilots installations
- central services: dataverse & triple store
*

requirements for semantics enabled machine learning
* lara intro
* lara intro
building homgeneous infrastructure from ground up
*
the greifswald protein screening platform LARA
* lara intro
SiLA servers/devices of LARA
*
demo
*
LARA, SiLA, AnIML/JSON-LD pythonLab, pythonLabScheduler, LabDataReader
*
pythonLab
https://gitlab.com/opensourcelab/pythonLab universal, python based, automation language
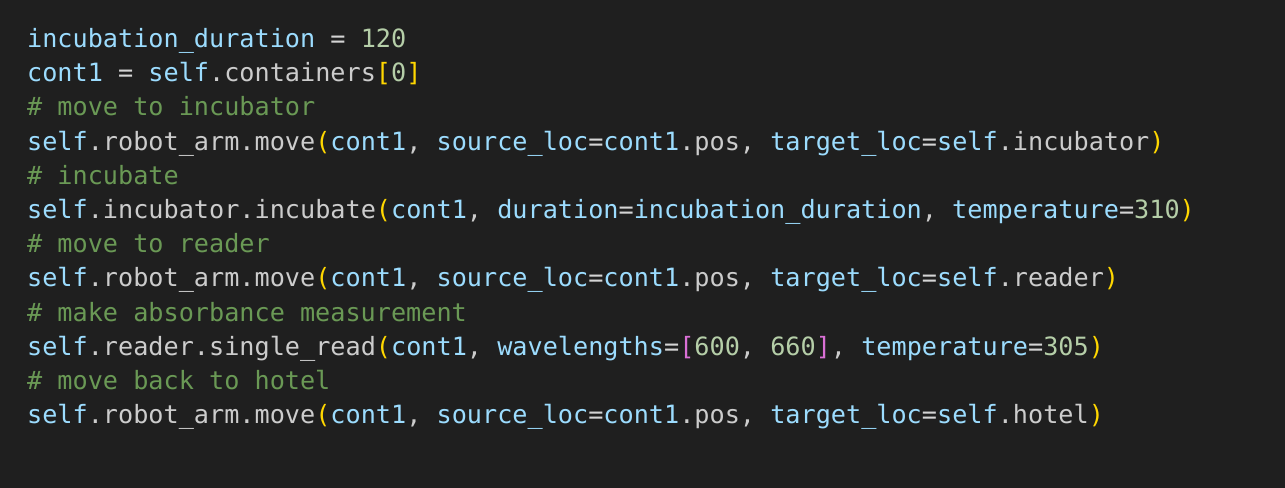
*
 pythonLabOrchestrator & Scheduler
pythonLabOrchestrator & Scheduler
https://gitlab.com/opensourcelab/pythonlabscheduler
*
 pythonLabScheduler in the wild (stefan maak)
pythonLabScheduler in the wild (stefan maak)
https://gitlab.com/opensourcelab/pythonlabscheduler
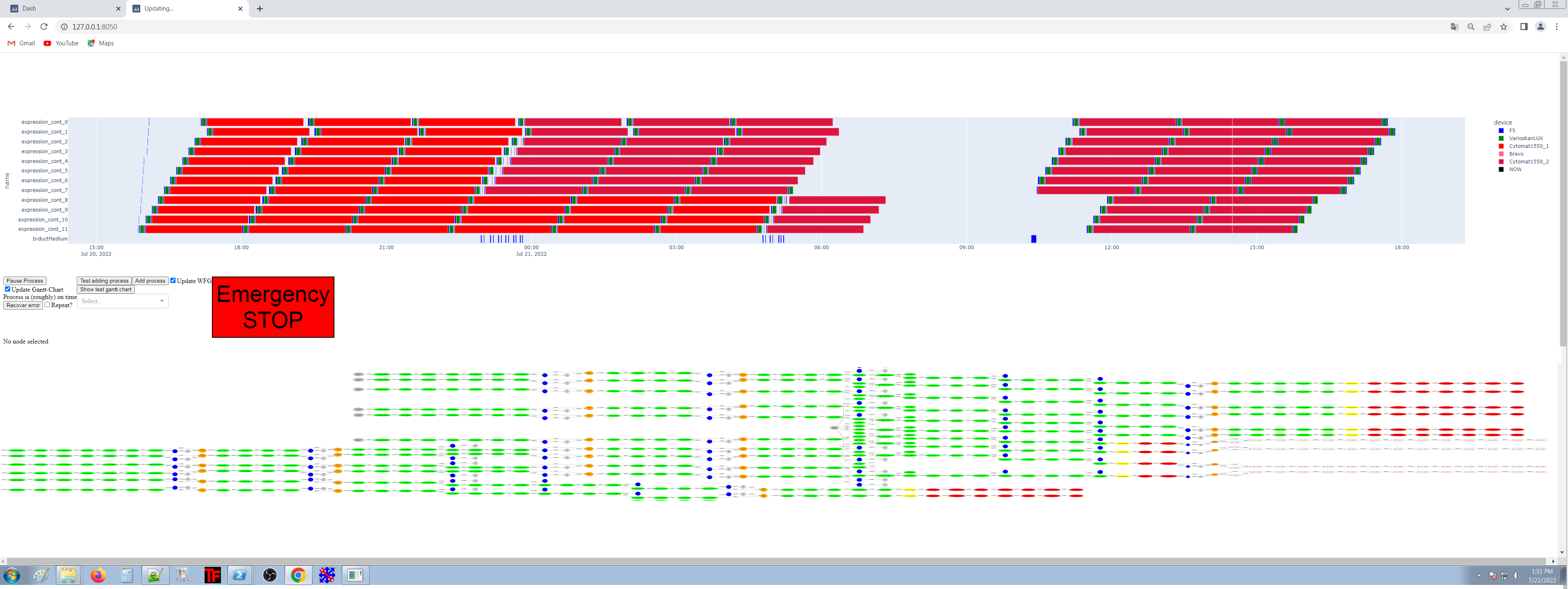
*
 LabDataReader
LabDataReader
https://gitlab.com/opensourcelab/LabDataReader
- generic data reader framework for propriatory (text based) data formats
- new data formats can be added as plugins
- rich meta data support
- automatic semantic annotation of the data
- fully written in python
- output formats: pandas data frame, JSON-LD, csv, (AnIML/JSON-LD - under development)
*
 holistic approach of the LARA suite
holistic approach of the LARA suite
- planning of experiments
- storing all required data for the planning, like literature, substances, material, devices, experimentalists ...
- generating the processes
- execution of the processes, communication with the lab devices
- collection of the data (very structured, well prepared to learn from it)
- evaluation and visualisation of the data (also DoE and machine learning)
- reporting / publishing / exchange between labs
* In the very early days of personal computing, I was wondering, why the computer was not used
 overview of final architecture
overview of final architecture
fully open sourced and python based
*

ontology - development - for semantic search / ML
*
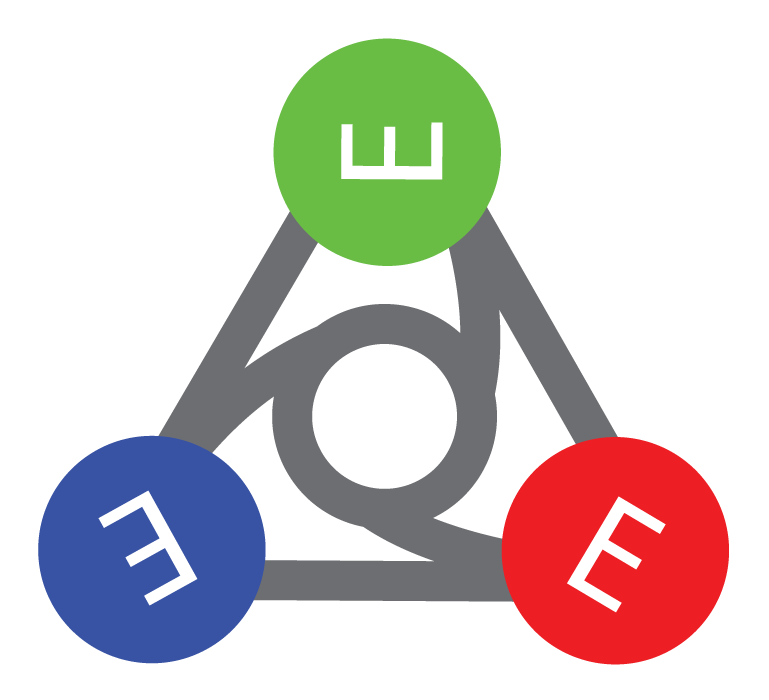 EMMO - European Multiperspective Material Ontology
EMMO - European Multiperspective Material Ontology
- top- & mid level ontology
- sould theoretical foundation
- rooted in historical philosopy (mereology), topology, physics and quantum physics
- all ITEMS are, e.g., SpaceTime Objects
- multiperspective and multidisciplinary
- modelling and experiments
- small -> fast reasoning
- python representation (EMMOntoPy)
*
 EMMOntoPy
(github.com/emmo-repo/EMMOntoPy)
EMMOntoPy
(github.com/emmo-repo/EMMOntoPy)
- all OWL classes and Properties are modeled as Python Objects
- generation of an ontology and reaoning can be done completely in python
- modular / object oriented modelling possible
- easy interaction / integration in own python applications
- SPARQL endpoint
- fast SQLITE triple store
- code managed in git repository
- tools for validation, documentation and visualisation
*
ontology development pipeline
*
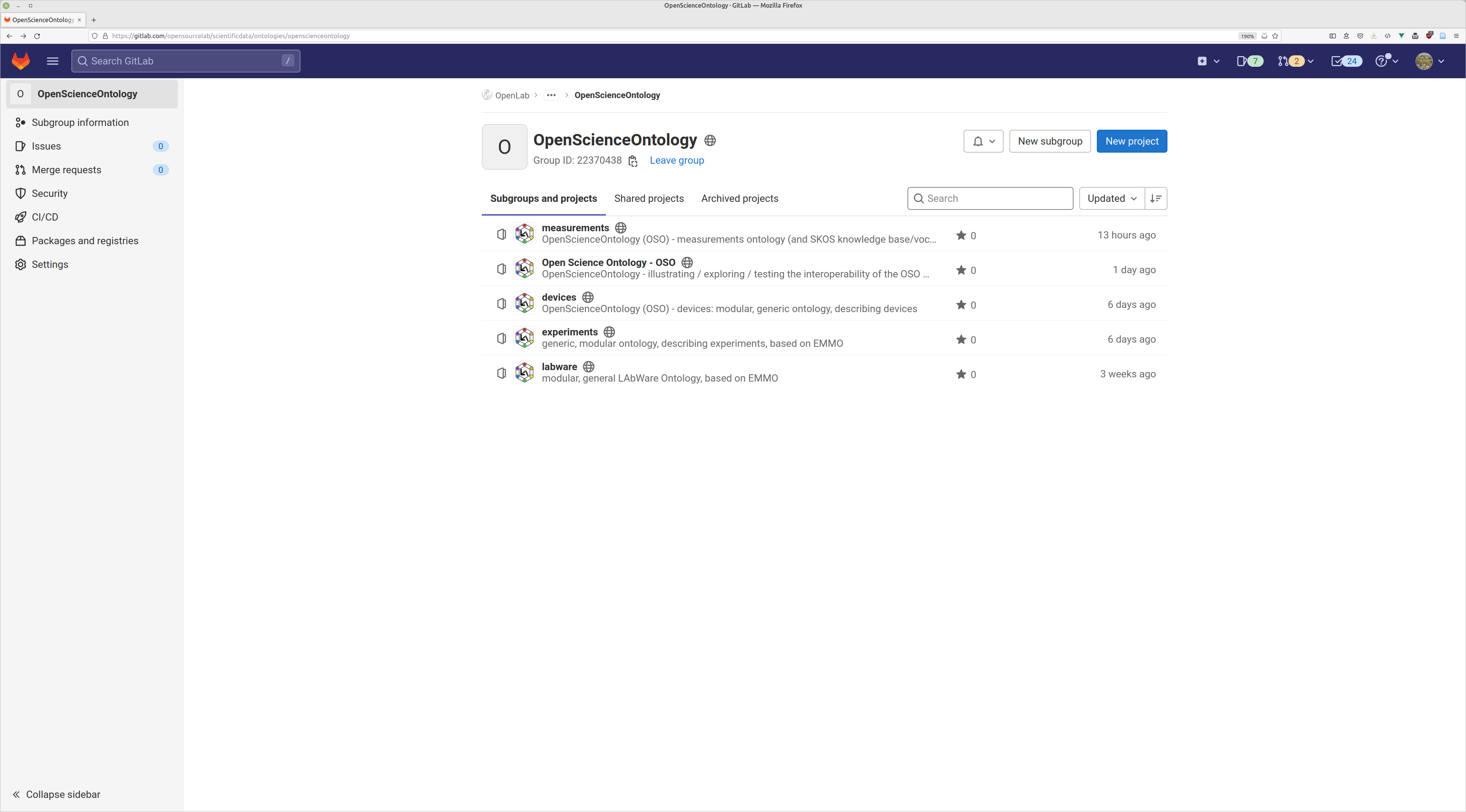
ontologies @ OpenSourceLab 
*
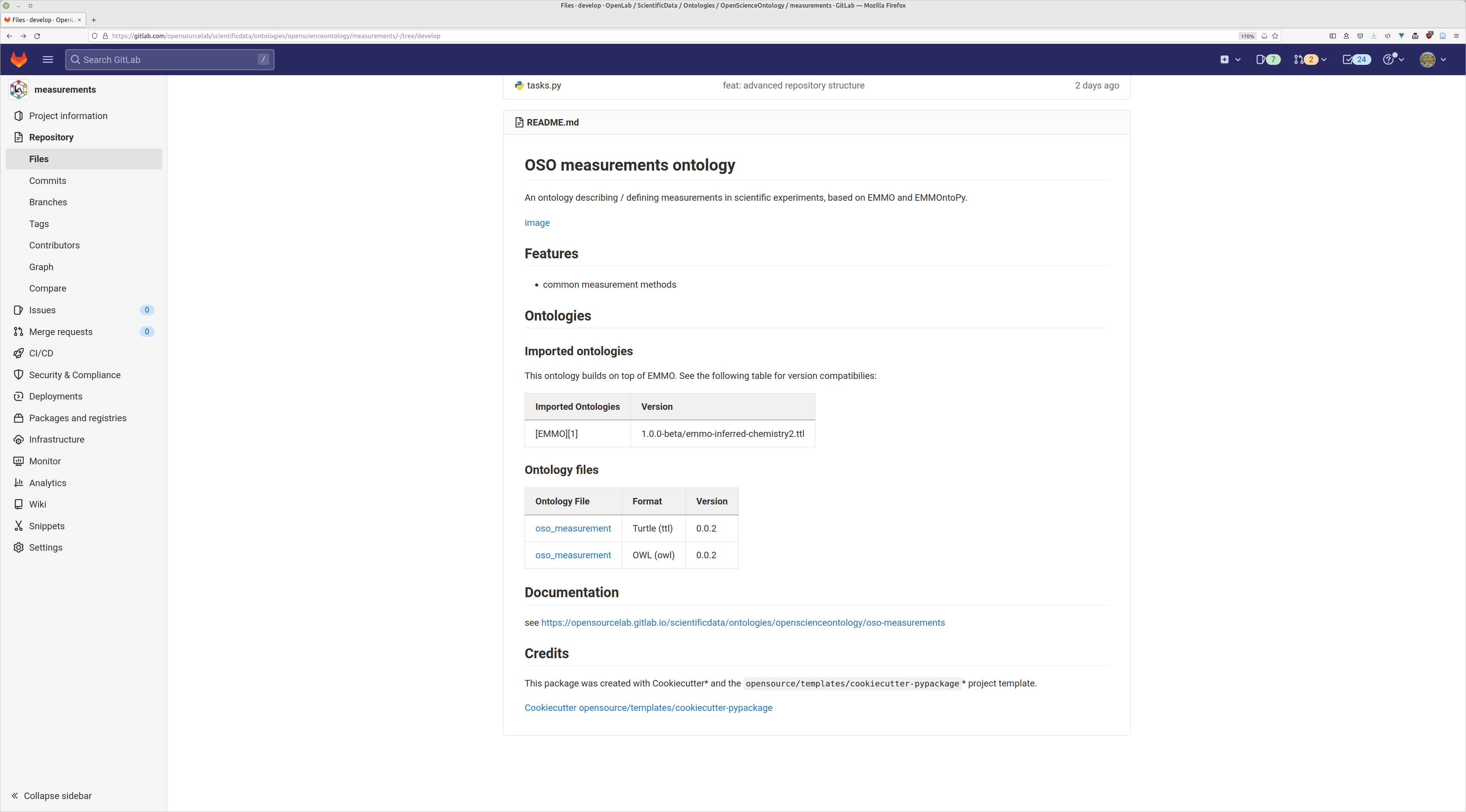
exmple: OSO measurement 
*

summary
- building a large, community driven open source infrastructure
- no black box, everything is adjustable and extendible
- advanced queries and automated feature extraction
- feedback loop driven protein engineering is possible with the tools in place
- machine learing applications will dramatically improve, when the right metadata is in place
- we need a modern mind set (biochemistry and machine learning)
- we are not alone, please join and contribute !
*
acknowledgements
project partners
- Stefan Born (TU Berlin)
- Peter Neubauer's group (TU Berlin)
- Johannes Kabisch's group and associates (Uni Trondheim)
- Egon Heuson (Uni Lille)
-
 Uwe Bornscheuer
and our group (Univ. Greifswald)
Uwe Bornscheuer
and our group (Univ. Greifswald)
KIWI-UG / NFDI4Cat
SiLA team
AnIML team
This work was supported by the German Federal Ministry of Education and Research through the Program “International Future Labs for Artificial Intelligence” (Grant number 01DD20002A)
We are grateful to the Deutsche Forschungsgemeinschaft (DFG, INST 292/ 118-1 FUGG) and the federal state Mecklenburg-Vorpommern for financing the robotic platform.
 National Research Data Infrastructure - for Catalysis
National Research Data Infrastructure - for Catalysis